FITExP or Functional Identification of Target by Expression Proteomics
- We can provide service for drug target finding based on compound-specific proteome responses and with no need of chemical engineering of compounds, using FITExP method.
- Target-based discovery is the way most pharmaceutical industry uses in searching for new drugs, with compound libraries screened for binding or activity against a known protein target.
- In contrast, phenomenological screening of small molecule libraries is a “black-box”, target-agnostic approach, where compounds are interrogated in cell-based assays with a readout linked to a disease-relevant process (e.g., cancer cell apoptosis).
- Arguably, this latter approach to drug discovery offers better chances for success. This is because the assay is more relevant to human physiology, and a multitude of targets are addressed simultaneously.
- It is known that, for the protein target of a small-molecule anti-cancer drug, the abundance change in late apoptosis is unexpectedly large compared to other proteins that are normally co-regulated with the drug target.
- FITExP method can help you to find one or several protein target(s) of your anti-cancer drug candidate from > 4000 proteins.
- Additionally, FITExP can unleash the mechanism of drug action (MoA) from the protein networks found in this methods.
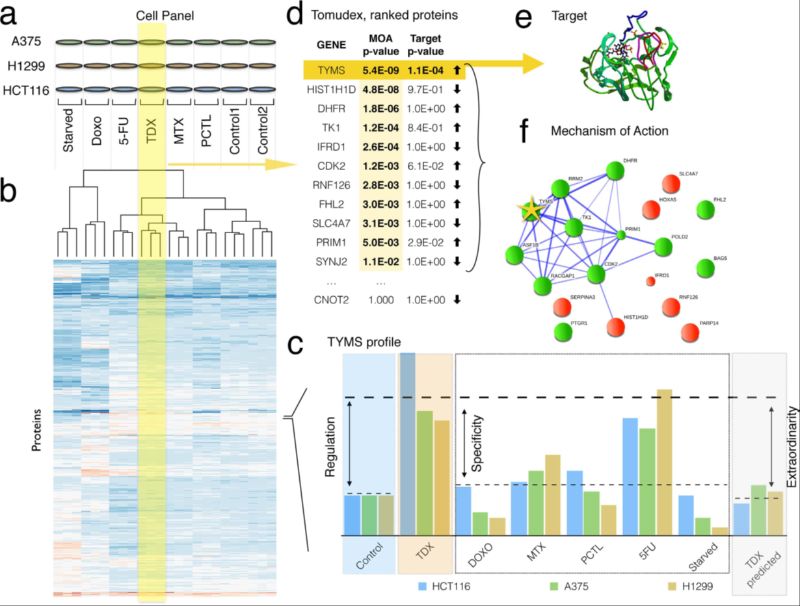
Figure. Example of workflow of the FITExP method of drug target identification: (a) a panel of cell lines is treated by a panel of drugs, in biological triplicates; (b) LC-MS/MS based proteomics identifies and quantifies ≥ 3,500 proteins, proteomic profiles are shown in a schematic heatmap with color-coded normalized abundances; the dendrogram shows hierarchical clustering of proteomic profiles with correlation-based distances; (c) for each protein, cell line and treatment, regulation (Reg), specificity (Spec) and exceptionality (Exc) are calculated; (d) for each treatment, final protein ranks based on Reg and Exc are established and the p-values are calculated using Bonferroni correction; protein list is sorted in ascending order of p-values; (e) few proteins with p ≤ 0.05 (threshold p-value) represent the most likely drug targets; (f) top n proteins with p ≤ 0.05 according to Reg and Spec rankings are mapped on protein networks to identify the drug target mechanism.
Reference:
- Scientific Reports (2015), 5:11176.
- Scientific Reports (2017), 7(1):1590.
- Proteomics (2018),18(24):e1800118.
- Redox Biol. (2020), 32: 101491.
- Redox Biol. (2021), 48: 102184.
Book a Meeting now to know how it can be beneficial / complement your R&D
